The three most common graphical models are Bayesian networks (aka directed acyclic graphs), Markov random fields (aka undirected graphs), and factor graphs.
Bayesian networks
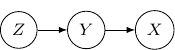
A Bayesian network describes the dependency of variables directly represented by the expansion of the conditional probabilities. Take a very simple example with three variables ![]() , and
, and ![]() . If the joint distribution can be represented by
. If the joint distribution can be represented by ![]() . Then, the corresponding Bayesian network will be a three-node graph with
. Then, the corresponding Bayesian network will be a three-node graph with ![]() , and
, and ![]() as vertices. And we have two directed edges one from
as vertices. And we have two directed edges one from ![]() to
to ![]() and another one from
and another one from ![]() to
to ![]() .
.
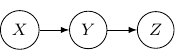
Note that by Bayes rule, the joint probability can be rewritten as ![]() . So the following Bayesian network with directed edges one from
. So the following Bayesian network with directed edges one from ![]() to
to ![]() and another one from
and another one from ![]() to
to ![]() is identical to the earlier one.
is identical to the earlier one.
Given ![]() , and
, and ![]() , we may also have an empty graph with no edge at all. This corresponds to the case when
, we may also have an empty graph with no edge at all. This corresponds to the case when ![]() that all three variables are independent as shown below.
that all three variables are independent as shown below.
![]()
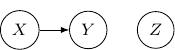
We may also have just one edge from ![]() to
to ![]() . This corresponds to the case when
. This corresponds to the case when ![]() , which describes the situation when
, which describes the situation when ![]() is independent of
is independent of ![]() and
and ![]() as shown below.
as shown below.
Now, back to the cases with two edges, the ones with two directed edges flowing the same direction will just have the same structure as the example we have earlier. So let’s look at other interesting setups. First, let’s consider the case that both directed edges are coming out of the same variable. For example, we have two directed edges from ![]() to
to ![]() and from
and from ![]() to
to ![]() as shown below.
as shown below.
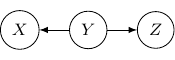
This corresponds to the joint distribution ![]() . So this case is actually the same as the case we have earlier after all. Note that however, this equivalence is not generalizable when we have more variables. For example, say we also have another variable
. So this case is actually the same as the case we have earlier after all. Note that however, this equivalence is not generalizable when we have more variables. For example, say we also have another variable ![]() and an edge from
and an edge from ![]() to
to ![]() .
.
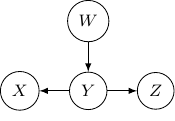
And if we have the direction of the edge ![]() to
to ![]() flipped as below,
flipped as below, ![]() and
and ![]() become independent but the earlier representation does not imply that.
become independent but the earlier representation does not imply that.
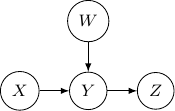
Now, let’s remove ![]() and assume we only have three variables and two edges as described earlier. For all cases we have above (say edges from
and assume we only have three variables and two edges as described earlier. For all cases we have above (say edges from ![]() to
to ![]() and
and ![]() to
to ![]() ), the variables at the two ends
), the variables at the two ends ![]() and
and ![]() are not independent. But they are independent given the variable
are not independent. But they are independent given the variable ![]() in the middle. Be very careful since the situation will be totally flipped as we change the direction of the edge from
in the middle. Be very careful since the situation will be totally flipped as we change the direction of the edge from ![]() to
to ![]() in the following.
in the following.
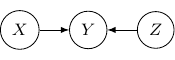
The corresponding joint probability for the above graph is ![]() . We have
. We have ![]() and
and ![]() are independent as
are independent as ![]() . On the other hand, we don’t have
. On the other hand, we don’t have ![]() . And so
. And so ![]() and
and ![]() are not conditional independent given
are not conditional independent given ![]() is observed. The classic example is
is observed. The classic example is ![]() and
and ![]() are two independent binary outcome taking values
are two independent binary outcome taking values ![]() and
and ![]() , where
, where ![]() is the xor operation such that
is the xor operation such that ![]() and
and ![]() . As the xor equation also implies
. As the xor equation also implies ![]() ,
, ![]() is completely determined by
is completely determined by ![]() when
when ![]() is given. Therefore,
is given. Therefore, ![]() is in no way to be independent of
is in no way to be independent of ![]() given
given ![]() . Quite the opposite,
. Quite the opposite, ![]() and
and ![]() are maximally correlated given
are maximally correlated given ![]() .
.
Let’s consider the case with three edges. Note that we can’t have edges forming a loop since Bayesian networks have to be an acyclic graph. A possibility can be having three directed edges one from ![]() to
to ![]() , one from
, one from ![]() to
to ![]() , and the last one from
, and the last one from ![]() to
to ![]() as shown below.
as shown below.
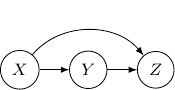
In this case, the joint probability will be ![]() . Note that the joint probability can be directly obtained by Bayes rule and so the graph itself does not assume any dependence/independence structure. The other Bayesian networks with three edges will all be identical and do not imply any independence among variables as well.
. Note that the joint probability can be directly obtained by Bayes rule and so the graph itself does not assume any dependence/independence structure. The other Bayesian networks with three edges will all be identical and do not imply any independence among variables as well.
As we see above, the dependency of variables can be derived from a Bayesian network as the joint probability of the variables can be written down directly. Actually, an important function of all graphical models is to represent such dependency among variables. In terms of Bayesian networks, there is a fancy term known as d-separation which basically refers to the fact that some variables are conditional independent from another given some other variables as can be observed from the Bayesian network. Rather than trying to memorize the rules of d-separation, I think it is much easier to understand the simple three variables examples and realize under what situations that variables are independent or conditionally independent. And for two variables to be conditionally independent of one another, all potential paths of dependency have to be broken down. For example, for the three variables three edges example above (with edges ![]() ,
, ![]() , and
, and ![]() ), observing
), observing ![]() will break the dependency path of
will break the dependency path of ![]() for
for ![]() and
and ![]() . But they are not yet conditionally independent because of the latter path
. But they are not yet conditionally independent because of the latter path ![]() .
.
Let’s consider one last example with four variables ![]() and
and ![]() , and three edges (
, and three edges (![]() ,
, ![]() , and
, and ![]() ) below.
) below.
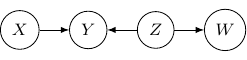
Note that as root nodes, ![]() and
and ![]() are independent. And thus
are independent. And thus ![]() and
and ![]() are independent as well since
are independent as well since ![]() only depends on
only depends on ![]() . On the other hand, if
. On the other hand, if ![]() is observed,
is observed, ![]() and
and ![]() are no longer independent. And thus
are no longer independent. And thus ![]() and
and ![]() are not conditionally independent given
are not conditionally independent given ![]() .
.
Undirected graphs and factor graphs
Compared with Bayesian networks, dependency between variables is much easier to understand. Two variables are independent if they are not connected and two variables are conditionally independent given some variables if the variables are not connected if the vertices of the given variables are removed from the graph.
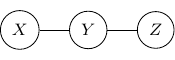
For example, consider a simple undirected graph above with three variables ![]() ,
, ![]() , and
, and ![]() with two edges
with two edges ![]() and
and ![]() . Variables
. Variables ![]() and
and ![]() are then independent given
are then independent given ![]() .
.
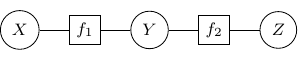
By Hammersley-Clifford theorem, the joint probability of any undirected graph models can be represented into the product of factor function of the form ![]() . For example, the joint probability of the above graph can be rewritten as
. For example, the joint probability of the above graph can be rewritten as ![]() with
with ![]() , and
, and ![]() . We can use a bipartite graph to represent the factor product above in the following.
. We can use a bipartite graph to represent the factor product above in the following.
- Represent each factor in the product with a vertex (typically known as factor node and display with a square by convention)
- Represent each random variable with a vertex (typically known as a variable node)
- Connect each factor node to all of its argument with an undirected edge
The graph constructed above is known as a factor graph. With the example described above, we have a factor graph as shown below.
The moralization of Bayesian networks
Consider again the simple Bayesian network with variables ![]() ,
, ![]() , and
, and ![]() , and edges
, and edges ![]() and
and ![]() below.
below.

What should be the corresponding undirected graph to represent this structure?
It is tempting to have the simple undirected graph before with edges ![]() and
and ![]() below.
below.

However, it does not correctly capture the dependency between ![]() and
and ![]() . Recall that when
. Recall that when ![]() is observed or given,
is observed or given, ![]() and
and ![]() are no longer independent. But for the undirected graph above, it exactly captured the opposite. That is
are no longer independent. But for the undirected graph above, it exactly captured the opposite. That is ![]() and
and ![]() are conditionally independent given
are conditionally independent given ![]() . So, to ensure that this conditional independence is not artificially augmented into the model, what we need to do is to add an additional edge
. So, to ensure that this conditional independence is not artificially augmented into the model, what we need to do is to add an additional edge ![]() as shown below.
as shown below.
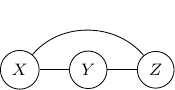
This procedure is sometimes known as moralization. It came for the analogy of a child born out of wedlock and the parents should get married and “moralized”.
Undirected graphs cannot represent all Bayesian networks
Despite the additional edge ![]() , the resulting graph, a fully connected graph with three vertices
, the resulting graph, a fully connected graph with three vertices ![]() ,
, ![]() , and
, and ![]() still does not accurately describe the original Bayesian network. Namely, in the original network,
still does not accurately describe the original Bayesian network. Namely, in the original network, ![]() and
and ![]() are independent but the undirected graph does not capture that.
are independent but the undirected graph does not capture that.
So if either way the model is not a perfect representation, why we bother to moralize the parents anyway? Note that for each edge we added, we reduce the (dependency) assumption behind the model. Solving a relaxed problem with fewer assumptions will not give us the wrong solution. Just maybe will make the problem more difficult to solve. But adding an incorrect assumption that does not exist in the original problem definitely can lead to a very wrong result. So the moralization step is essential when we convert directed to undirected graphs. Make sure you keep all parents in wedlock.
Bayesian networks cannot represent all undirected graphs
Then, does that mean that Bayesian networks are more powerful or have a higher representation power than undirected graphs? Not really also, as there are undirected graphs that cannot be captured by Bayesian networks as well.
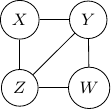
Consider undirected graph with four variables ![]() and
and ![]() and four edges
and four edges ![]() ,
, ![]() ,
, ![]() , and
, and ![]() as shown below.
as shown below.
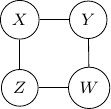
Note that we have conditional independence of ![]() and
and ![]() given
given ![]() and
and ![]() . This conditional independence is captured only by one of the following three Bayesian networks.
. This conditional independence is captured only by one of the following three Bayesian networks.
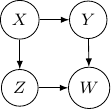
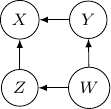
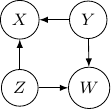
Because of the moralization we discussed earlier, when we convert the above models into undirected graphs, they all require us to add an additional edge between parents ![]() and
and ![]() resulting in the undirected graph below.
resulting in the undirected graph below.
Note that this undirected graph is definitely not the same as the earlier one since it cannot capture the conditional independence between ![]() and
and ![]() given
given ![]() and
and ![]() . In conclusion, no Bayesian network can exactly capture the structure of the square undirected graph above.
. In conclusion, no Bayesian network can exactly capture the structure of the square undirected graph above.